1.What is the 3CLP??
3CLP is an online tool for predicting coronavirus 3CL protease cleavage sites.It also predicted human protein sequences from the Swiss-prot database.Potential cleavage sites on human protein are available for query and download in Data interface.Training data and testing data for 3CLP is available for download.
2.How to use the 3CLP?
- User input the protein sequence(either coronavirus polyprotein,or the host proteins) in Fasta format,the output is the potential cleavage sites and the corresponding score predicted by 3CLP.
- Before using 3CLP, please ensure your protein sequence is in Fasta format and “-” is not allowed in the sequence.Your protein ID should not contain special symbol *.
- Don’t input more than 100 protein sequences per time.
- In the prediction result interface,it only presents the cleavage sites which score is more than 0.5.All predicted results were available for download.The prediction result file contains your protein name,position of Q(Gln),cleavage motif and its score.
- If the prediction result in the result interface is empty,it may be because 3CLP can not obtain valid test sample from your protein sequence or the score of all test sample are less than 0.5.Your can download the result file to check all of your prediction result.
3.What is the coronavirus 3CL protease?
- The 3C-like (3CL) protease is a Cysteine protease. As a major viral protease, 3CL protease usually specifically recognize the 11 cleavage sites of nsp4~nsp16 in pp1ab and these nonstructural proteins released by cleavage participate in viral genome replication and transcription.
- The cleavage sites of coronavirus 3CL protease have a feature.The first position in the upstream of the cleavage site,defined as P1 position,was highly conserved with the amino acid(AA) Q(Gln).Therefore,3CLP only consider whether there are potential cleavage sites near the Q(Gln) in the protein sequence.If the protein sequence provided by user does not contain Q(Gln),the prediction result of 3CLP is empty.
4.What is the known cleavage site?
- We found mature peptide annotations of 14 coronavirus species in three genera including Alphacoronavirus, Betacoronavirus and Gammacoronavirus from the NCBI RefSeq and protein database.It is known that coronavirus 3CL protease specifically recognize the 11 cleavage sites of nsp4~nsp16 in pp1ab,therefore the known cleavage sites are between these nonstructural proteins.The known cleavage sites of 14 coronavirus species are available for query in Data interface.
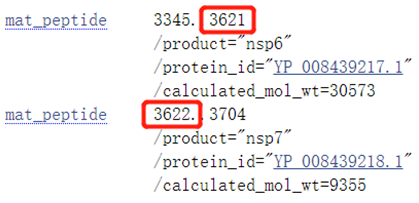
- For example,as shown below,the last amino acid(AA) of nsp6 is located at 3621 position and the first amino acid(AA) of nsp7 is located at 3622 position.The known cleavage site is between the 3621 position and the 3622 position.The motif near the cleavage site is STVQ↓SK.(The Screenshot is from NCBI)
5.How to build the RF model of predicting the cleavage sites?
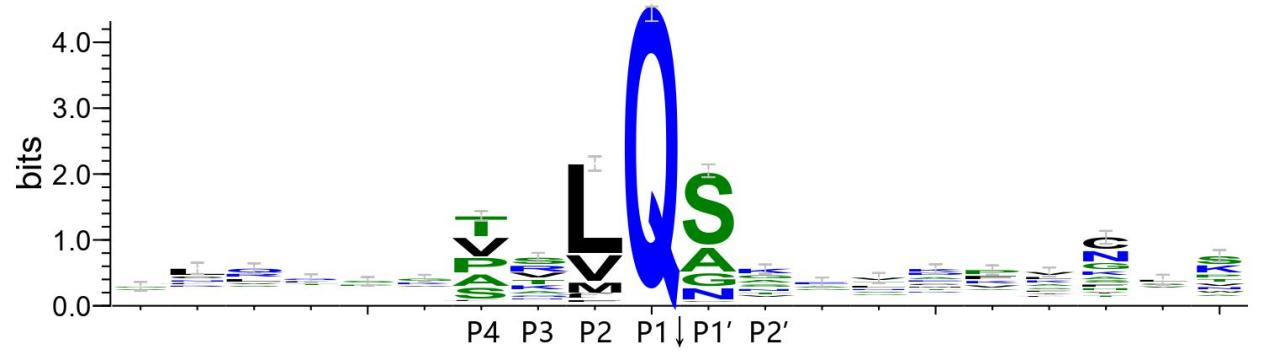
- Cleavage motif:The logos of sequences around the cleavage sites for three genera (14 coronavirus species) showed the P1-P4 and P1'-P2' were more conserved than other positions.Therefore these 6 positions defined as the cleavage motif.
- Trainset:The train data for 3CLP contains 265 cleavage motifs (positive samples) and equal number of non-cleavage motifs (negative samples).
- Feature:Each amino acid(AA) on the positive or negative sample (the cleavage or non-cleavage motif) was encoded by 3 AA indexes of of MEEJ800102, BIOV880102 and FASG760101, which referred to “the retention coefficient in high-pressure liquid chromatography”, “Information value for accessibility” and “Molecular weight”, respectively. (AA indexes are from the AAindex database).
- Model:We used the RF(random forest) model for predicting the cleavage sites of the coronavirus 3CL protease.The AUC,specifically,accuracy,sensitivity, FPR of the model were 0.96, 0.88, 0.80, and 0.04, respectively.
- For more details about the prediction method, please refer to our study.
6.How to obtain the cleavage sites on human proteins?
- The human proteome was obtained from the SwissProt database in UniProt.
- 20386 human proteins were predicted by 3CLP. A total of 1352 human proteins were predicted to be cleaved by the coronavirus 3CL protease with 1511 cleavage sites.
- All 1511 cleavage sites on human proteins are available for query in Data interface.
- All potential cleavage sites(taking 0.5 as cutoff) on human proteins are available for download in Data interface.
7.How to cite us ?
If you use the 3CLP, please cite our manuscript as follows:
Huiting Chen, Zhaozhong Zhu, Yousong Peng. Prediction of coronavirus 3C-like protease cleavage sites using machine-learning algorithms. Manuscript in preparation.